| Academic Appointments
2014 - Present Principal Investigator. Institute of Genetics and Developmental Biology, Chinese Academy of Sciences
2013 - 2014 Member of the Professional Staff, California Institute of Technology. Approved by the Institute Academic Council (IACC) |
2010 - 2013 Senior Research Fellow (Voting Faculty), California Institute of Technology. Approved by IACC
2004 - 2010 Postdoctoral Scholar, California Institute of Technology.
2003 - 2004 Research Associate, Research Center for Proteome Analysis, Chinese Academy of Sciences
Education
2003 Ph.D. Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences
2000 M.Sc. Fudan University
1997 B.Sc. Anhui Normal University
Research interest
Our research interest is in developmental systems biology. We seek to understand the big picture of how all the components interact in complex developmental biology systems. More specifically, our research has focused on the study of gene regulatory networks (GRNs) that underlie development, regeneration, and diseases. Why do we study GRNs? Because a single gene cannot dictate a biological process, no matter how important it is. A regulatory mechanism is not just about one gene; it is about all genes; It is not just about genes, it is about causal linkages.
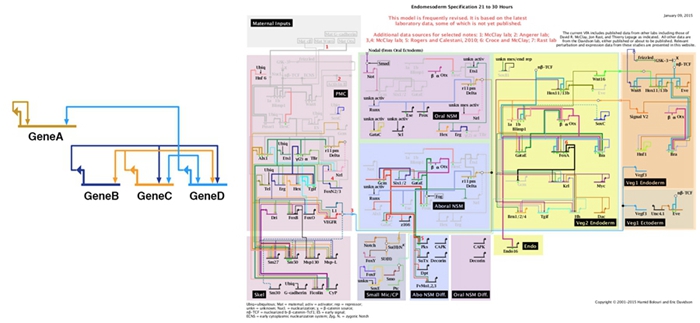
We employ a strategy that integrates both experimental and computational approaches, including genomics, epigenomics, single-cell technologies, gene perturbation, and bioinformatics. We identify the relevant genes, profile their activities, perturb their functions, quantitatively measure the causal changes, and integrate all the information into knowledge. We also develop novel tools essential for our research. Currently, we are trying to elucidate the regulatory programs governing sex determination, differentiation, and germ cell development.
The model organisms used in the lab are zebrafish and medaka. The zebrafish (Danio rerio) is one of the most popular model organism in life science. This tiny tropical fish is widely used in many research fields because of its easy maintenance, breeding, and transparent body during early development. Medaka (Oryzias latipes) is also a small freshwater fish and is native to East Asia. We have found wild medaka in the Olympic Forest Park near the lab. Compared to zebrafish, medaka has many unique characteristics, both genetic and physiological. Medaka lives in small rivers, creeks and rice paddies, and can stand a wide range of temperatures (4 - 40℃) and salinities. Additionally, medaka has clearly defined sex chromosomes, and its sex determination mechanism has been intensively studied over the years. It has become an important vertebrate model widely used in genetics, developmental biology, endocrinology, biomedical science, and environmental sciences.
Selected Publications
- Wang F*, Jia W*, Fan M*, Shao X*, Li Z, Liu Y, Ma Y, Li Y-X, Li R#, Tu Q#, Wang Y-L#. 2021. Single-cell Immune Landscape of Human Recurrent Miscarriage. Genomics Proteomics Bioinformatics. [DOI]
- Li Y*, Liu Y*, Yang H, Zhang T, Naruse K#, Tu Q#. 2020. Dynamic transcriptional and chromatin accessibility landscape of medaka embryogenesis. Genome Res 30: 924–937. [DOI | Website]
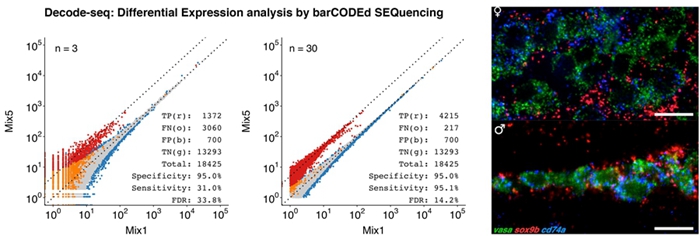
- Li Y*, Yang H, Zhang H, Liu Y, Shang H, Zhao H, Zhang T, Tu Q#. 2020. Decode-seq: a practical approach to improve differential gene expression analysis. Genome Biol 21: 66. [DOI | Website]
- Barsi JC*, Tu Q*, Calestani C, Davidson EH. 2015. Genome-wide assessment of differential effector gene use in embryogenesis. Development 142: 3892–3901. [ PDF | Website ]
- Barsi JC*, Tu Q*, Davidson EH. 2014. General approach for in vivo recovery of cell type-specific effector gene sets. Genome Res 24: 860–868. [ PDF | Website ]
- Tu Q, Cameron RA, Davidson EH. 2014. Quantitative developmental transcriptomes of the sea urchin Strongylocentrotus purpuratus. Dev Biol 385: 160-167. [ PDF | Website]
- Tu Q, Cameron RA, Worley KC, Gibbs RA, Davidson EH. 2012. Gene structure in the sea urchin Strongylocentrotus purpuratus based on transcriptome analysis. Genome Res 22: 2079–2087. [ PDF | Website ]
- Oliveri P*, Tu Q*, Davidson EH. 2008. Global regulatory logic for specification of an embryonic cell lineage. Proc Natl Acad Sci USA 105: 5955-5962. [ PDF | PNAS Feature Article| PNAS Comment | F1000 Comment ]
- Tu Q, Brown CT, Davidson EH, Oliveri P. 2006. Sea urchin Forkhead gene family: Phylogeny and embryonic expression. Dev Biol 300: 49-62. [ PDF ]
- Howard-Ashby M, Materna SC, Brown CT, Tu Q, Oliveri P, Cameron RA, Davidson EH. 2006. High regulatory gene use in sea urchin embryogenesis: Implications for bilaterian development and evolution. Dev Biol 300: 27-34.
- Samanta MP, Tongprasit W, Istrail S, Cameron RA, Tu Q, Davidson EH, Stolc V. 2006. The transcriptome of the sea urchin embryo. Science 314: 960-962.
- Tu Q, Tang H, Ding D. 2004. MedBlast: searching articles related to a biological sequence. Bioinformatics 20: 75-77.
- Tu Q, Ding D. 2003. Detecting pathogenicity islands and anomalous gene clusters by iterative discriminant analysis. FEMS Microbiol Lett 221: 269-275.
- Tu Q, Yu L, Zhang P, Zhang M, Zhang H, Jiang J, Chen C, Zhao S. 2000. Cloning, characterization and mapping of the human ATP5E gene, identification of pseudogene ATP5EP1, and definition of the ATP5E motif. Biochem J 347: 17-21.
- Zheng L, Yu L, Tu Q, Zhang M, He H, Chen W, Gao J, Yu J, Wu Q, Zhao S. 2000. Cloning and mapping of human PKIB and PKIG, and comparison of tissue expression patterns of three members of the protein kinase inhibitor family, including PKIA. Biochem J 349: 403-407.
- Lang T, Yu L, Tu Q, Jiang J, Chen Z, Xin Y, Liu G, Zhao S. 2000. Molecular cloning, genomic organization, and mapping of PRKAG2, a heart abundant gamma2 subunit of 5'-AMP- activated protein kinase, to human chromosome 7q36. Genomics 70: 258-263.
- 屠强. 1997. 植物分类学工具软件 (PTT) 的设计. 安徽师大学报 (自然科学版) 20: 189-193.
|



